|
科学哲学ニューズレター |
![]()
No. 40, April 3, 2001
Soshichi Uchii
The Human Genome Project: from Nature 409
Editor: Soshichi Uchii
The famous science journal Nature recently published a special issue concentrating on the "human genome project (HGP)" (409, 813-969,15 Feb. 2001). Since this contains many educational materials (including a CD-ROM), let me skim some of the essential features off this issue. But, first, I must warn the reader that I am no expert in this field; I am writing only in the capacity of a philosopher of science, and a teacher, interested in this field. Japanese readers may refer to Matsubara & Nakamura 1996 (in Japanese), for a popular exposition of the project at an early stage of the research in Japan.
Now, in the initial "News and Views" section, David Baltimore begins his overview "Our genome unveiled" (814-816), as follows:
I've seen a lot of exciting biology emerge over the past 40 years. But chills still ran down my spine when I first read the paper that describes the outline of our genome and now appears on page 860 of this issue. ... it is a seminal paper, launching the era of post-genomic science.But what is so exciting about HGP? First of all, as you may already know, HGP is a megaproject involving and coordinating 20 laboratories and hundreds of researchers around the world. Actually, this is true only of the public project, International Human Genome Sequencing Consortium; there is another project by Celera Genomics (a company), which takes advantage of the public project (its results are available to anyone) but intends to make money on the results obtained. But, anyway, the second exciting feature is that scientists all over the world are collaborating, not necessarily competing for priority, with each other. Moreover, the project aims at grasping the whole of human genome, not at analyzing single genes or any particular parts contained in it; thus it is one of the rare instances of synthesizing or integrating project. Finally, since this big project is bound to have important implications for human life and society, the ethical issues are explicitly tackled, and included as an indispensable part of the project.
Now the project launched in 1990 (in the US, and then many countries joined), and has produced, as a conspicuous result, "draft sequences" of human genome (one of the goals envisaged by the1998 HGP new 5 year plan), and that's the reason why Nature presented a special issue. According to Baltimore,
The most exciting new vista to come from the human genome is not tackling the question "What makes us human?", but addressing a different one: "What differentiates one organism from another?". The first question, imprecise as it is, cannot be answered by staring at a genome. The second, however, can be answered this way because our differences from plants, worms and flies are mainly a consequence of our genetic endowments.This may give you a glimpse of HGP. Making draft sequences is not confined to human genome; bacteria, archaea, and multicellular organisms such as a fly and a mouse are also subject (see, for instance, p. 819 of Nature 409, and the timeline in the attached CD-ROM). Further, as Maynard Olson comments, "This issue of Nature celebrates a halfway point in the implementation of the 'map first, sequence later' strategy adopted by the Human Genome Projct in the mid-1980s" (816). But many readers may wonder, "what's map? what's sequence?" Thus we have to review at least some of the major events leading to HGP, and we have to understand the basis of HGP, as well as its accomplishments so far.
Major events leading to the human genome project
1909 The word "gene" was created by a Danish botanist. Wilhelm Johannsen used this word in order to refer to Mendelian units of heredity. He also distinguished "genotype" from "phenotype".
1911 T. H. Morgan and his group contributed greatly to showing that genes are caried by chromosomes and genes are indeed units of heredity. They used the fruit fly, Drosophila melanogaster, for their research.
1952 Alfred Hershey and Martha Chase established, on the experimental basis, the idea that "genes are carried by DNA (in chromosomes)".
1953 Francis Crick and James Watson found the "double helix" structure of DNA. Together with M. Wilkinson who contributed to clarifying the same structure by means of X-ray diffraction patterns, they received the Nobel Prize (physiology) in 1962.
1955 Joe Hin Tjio made it clear that there are 46 human chromosomes (i.e. 23 pairs.)
[Tjio has a very unusual career. He was born in 1919 in Java, then the Dutch West Indies; he is a Chinese. During the war, he was imprisoned and spent three years in a concentration camp of Japanese Army. After the war, he obtained an opportunity to study in Europe; then he made research in Sweden and Denmark, and his discovery was made in Lund, Sweden. Herman Muller persuaded Tjio to come to the United States, and Tjio eventually continued his research in National Institute of Health (NIH). For more on Tjio, see http://www.nih.gov/news/NIH-Record/02_11_97/story01.htm]1961 Messenger RNA (mRNA) was discovered.
1966 Genetic code was deciphered; a triplet (of A, C, G, and T) encodes an amino acid necessary for organisms. Thus DNA may be regarded as a program of "how to make amino acids".
1972 Stanley Cohen and Herbert Boyer discovered the technique of Recombinant DNA.
1975-77 DNA sequencing method was established. This enables one to read DNA "letter by letter" (A, C, G, and T), very quickly.
1977 Introns are discovered; genes on DNA are often interrupted by chunks which do not carry genes, and these chunks are called introns.
[When mRNA transfers genetic code in order to make proteins, introns are omitted and genes are copied on mRNA; utilizing this property, mRNA can be copied back to DNA, called cDNA (complimentary DNA) without introns. Thus "cDNA library" is useful for studying genes.]1983 Huntington's disease is linked to a gene on chromosome 4; this is the first disease gene (and it bacame a "marker" of genetic map; many other markers are found subsequently, and they serve as a marker because DNA is different from others at that location depending on individuals).
[Maybe a note on "marker" is in order here. A marker is a segment of DNA with an identifiable location on a chromosome; and its inheritance must be recognized. A marker is not necessarily a gene; it may be any segment with, or without, a known function.]Polymerase chain reaction (PCR) method was discovered, and it enabled researchers to produce many copies of any DNA segment.
1987 The first human genetic map; this makes use of various polymorphisms (those parts of DNA which vary among people, and these can be used as markers).
YACs, that is, yeast artificial chromosomes, enabled to store and reproduce a large segment of DNA of other species, of humans in particular. [This enables researchers to make "libraries" of DNA segments. This technique is extended later as BACs, mainly utilizing Escherichia coli (bacterium), E. coli for short.]
1990 The launch of Human Genome Project, HGP. In the US, National Institute of Health (NIH) and Department of Energy (DOA) published a plan for the first 5 years (of the whole 15-year project). Also, Ethical, Legal and Social Implications (ELSI) programs were founded at NIH and DOE.
1996 The first international strategy meeting on human genome sequencing.
1997 UNESCO issued "UNIVERSAL DECLARATION ON THE HUMAN GENOME AND HUMAN RIGHTS" [see http://www.unesco.org/human_rights/hrbc.htm]
1998 Celera Genomics company announced to begin sequencing human genome, by a different method from that of HGP. They intend to utilize their results on commercial basis.
New HGP goals for 2003 published.
[For a more detailed timeline, see the CD-ROM attached to the special issue; it has movies and many illustrations which are quite useful.]
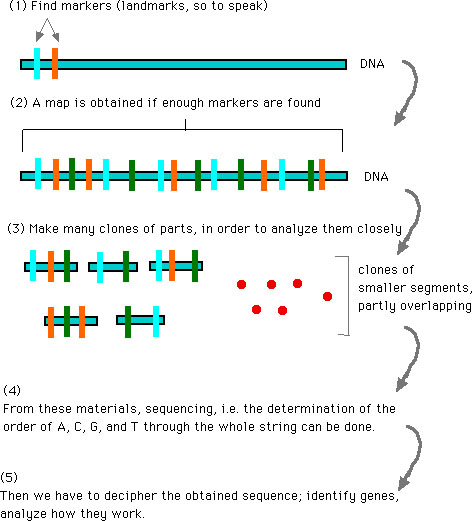
All right. With the preceding information, maybe you can begin to have a rough idea how the project goes. So let me show you a schematic view of DNA sequencing (adapted from a movie in the CD-ROM), which is nearly completed for human genome and gave "draft sequences", referred to in the preceding.
How the Sequencing Project goes: (4) and (5) are expected by 2003
The impact of this project on human life is expected to be immense. Let me just mention its implications for medical science. In the concluding part of the paper "Implications of the Genome project for Medical Science" (in the CD-ROM), the authors, Francis Collins and others, say as follows:
As we cross the threshold of the new millenium, we simultaneously cross a threshold into an era where the human genome sequence is largely known. We must commit ourselves to exploring the application of these powerful tools to the alleviation of human suffering, a mandate that undergirds all of medicine. At the same time, we must be mindful of the great potential for misunderstanding of a field that is developing very quickly, and make sure to advance the ethical and legal consideration of genetics with just as much vigor as the medical research.Scientific researches, especially those directly concerned with ourselves, have both good and (often unexpected) bad effects. But maybe the last statement of the quoted passage is too mild. We already have bitter experiences with "eugenics", nuclear weapons, and various kinds of chemicals; and we can easily imagine "undesirable" impact of HGP on human life, to wit, the problems of privacy, of discrimination based on genetic traits, of the combination of its knowledge with "genetic engineering" (which enables another sort of "eugenics" on individual and commercial basis). That's why ELSI was founded at the beginning of HGP, and UNESCO responded quickly by means of "Universal Declaration". If it becomes fashionable to talk about "Information Ethics", the problem of "genetic information" and its ethical problems should certainly be included as one of the "top priorities". Some of my colleagues in Kyoto University have published two volumes on the HGP and society (Kato 1995, 1996), but nearly two fifths are abstracts or summaries of Western literature, and their contents are not very impressive.
I must emphasize here that in order to make any positive contributions to solving these problems, philosophers must know at least essential features of the current research going on in the HGP. This means that they must read this issue of Nature, for instance, and prepare their groundwork for their own job. Practicing scientists, on the other hand, must become familiar with the ways for handling these ethical issues, if they seriously want to discuss them; otherwise their opinions as regards these issues will remain "mere opinions" or "mere intuitions". If they misunderstand (arrogantly) that an expert in their own field is also an expert in ethical issues, they are bound to repeat similar errors as those committed by some eminent scientists in the past. Philosophers and scientists both need a common ground, and presumably a mere "division of labor" will not work. But don't misunderstand what I am saying; I am not urging expertise in both fields.
The situation may be likened to the way for handling many clones overlapping with each other (see the preceding Figure); only with an appropriate overlapping, one can be sure that two segments are consecutive and become a larger part of the whole. I am suggesting similar things (no doubt a vague metaphor as it stands), as regards collaboration between scientists and philosophers, or anyone else. My own example may not be a good example, but let me say this much: I myself have been trying to fulfill my own part in preparing this sort of common grounds between philosophers on the one hand, and scientists such as physicists and biologists on the other.
Reference
Matsubara & Nakamura (1996) 松原謙一・中村桂子『ゲノムを読む』紀伊國屋書店。
Kato, H., ed. (1995) 『ヒトゲノム解析研究と社会との接点、研究報告集』、京都大学文学部倫理学研究室、1995。
Kato, H., ed. (1996) 『ヒトゲノム解析研究と社会との接点、研究報告集、第2集』、京都大学文学部倫理学研究室、1996。
Last modified Dec. 1, 2008. (c) Soshichi Uchii